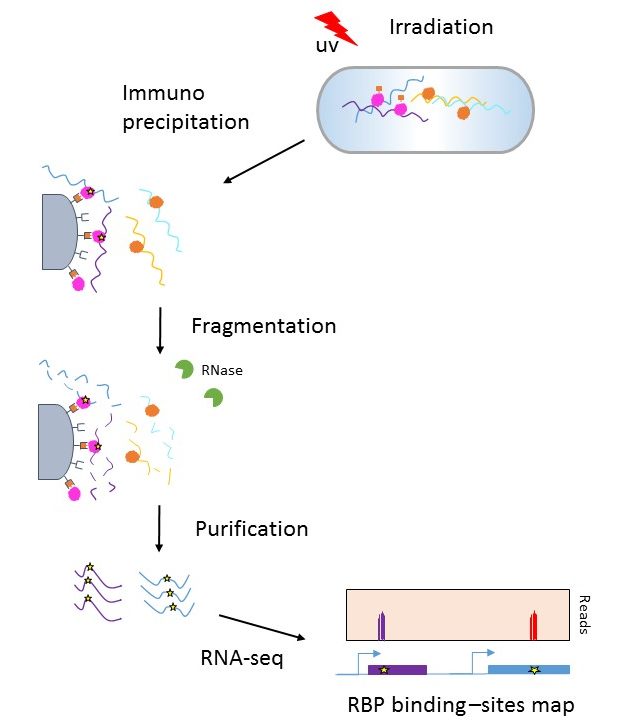
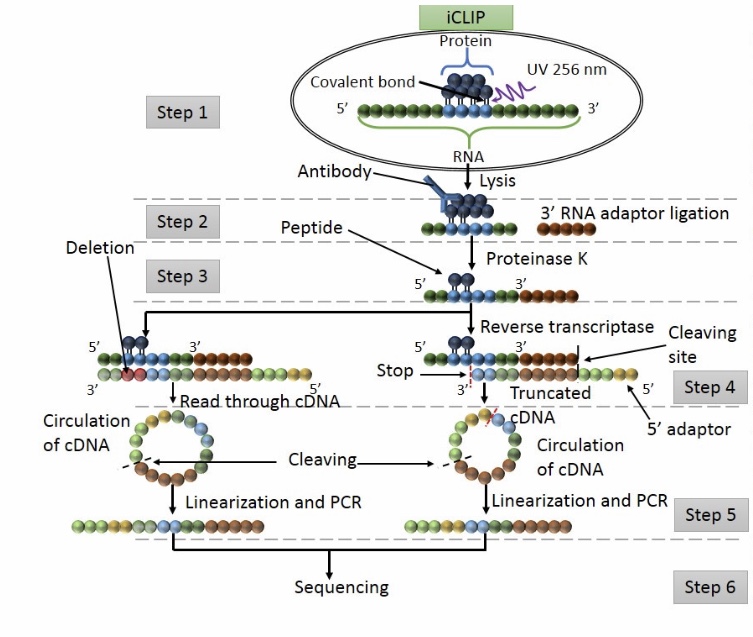
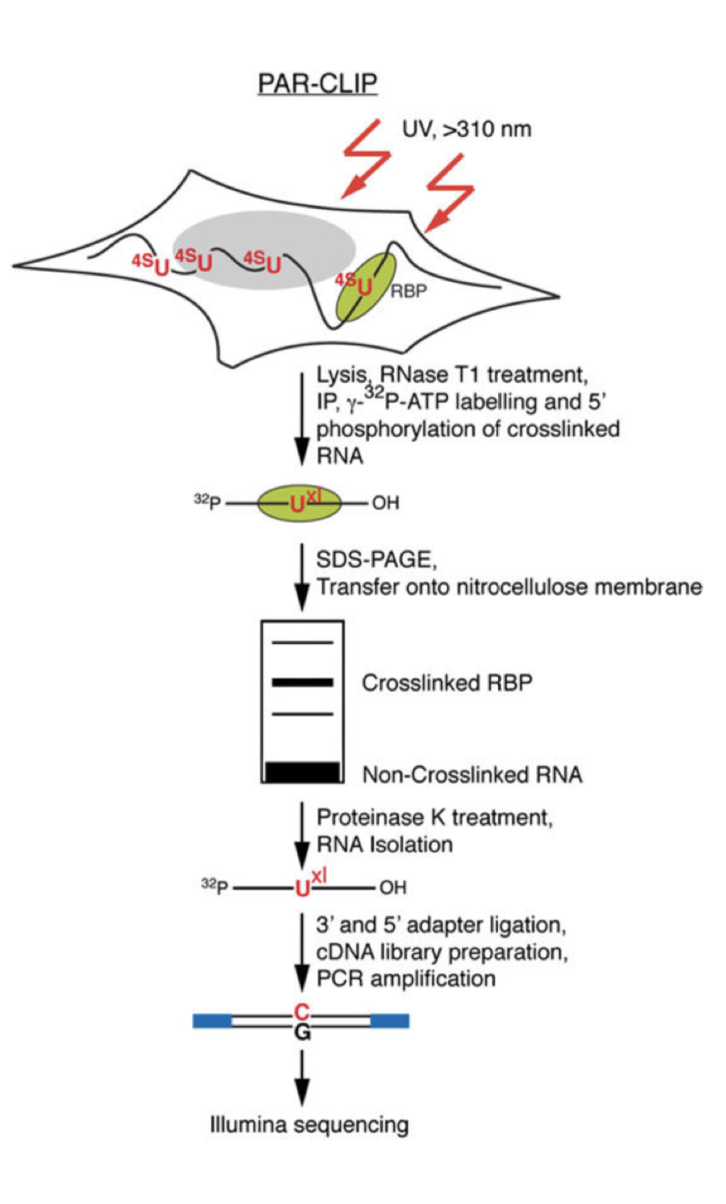
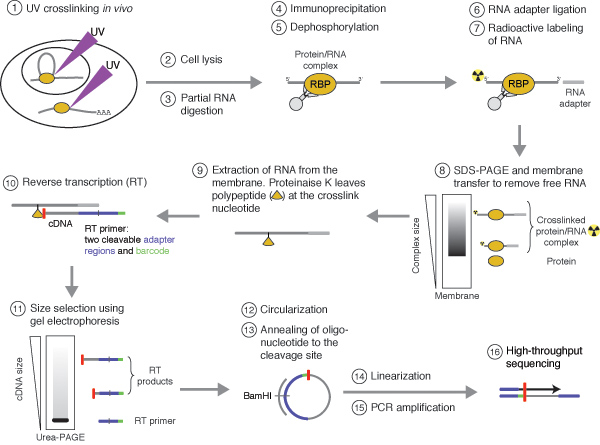
Figure 1 | PAR-CLIP: A Method for Transcriptome-Wide Identification of RNA Binding Protein Interaction Sites | SpringerLink

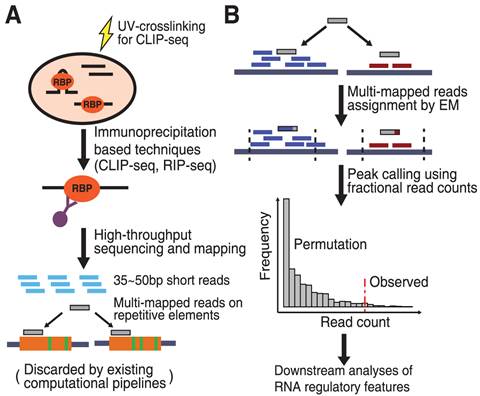
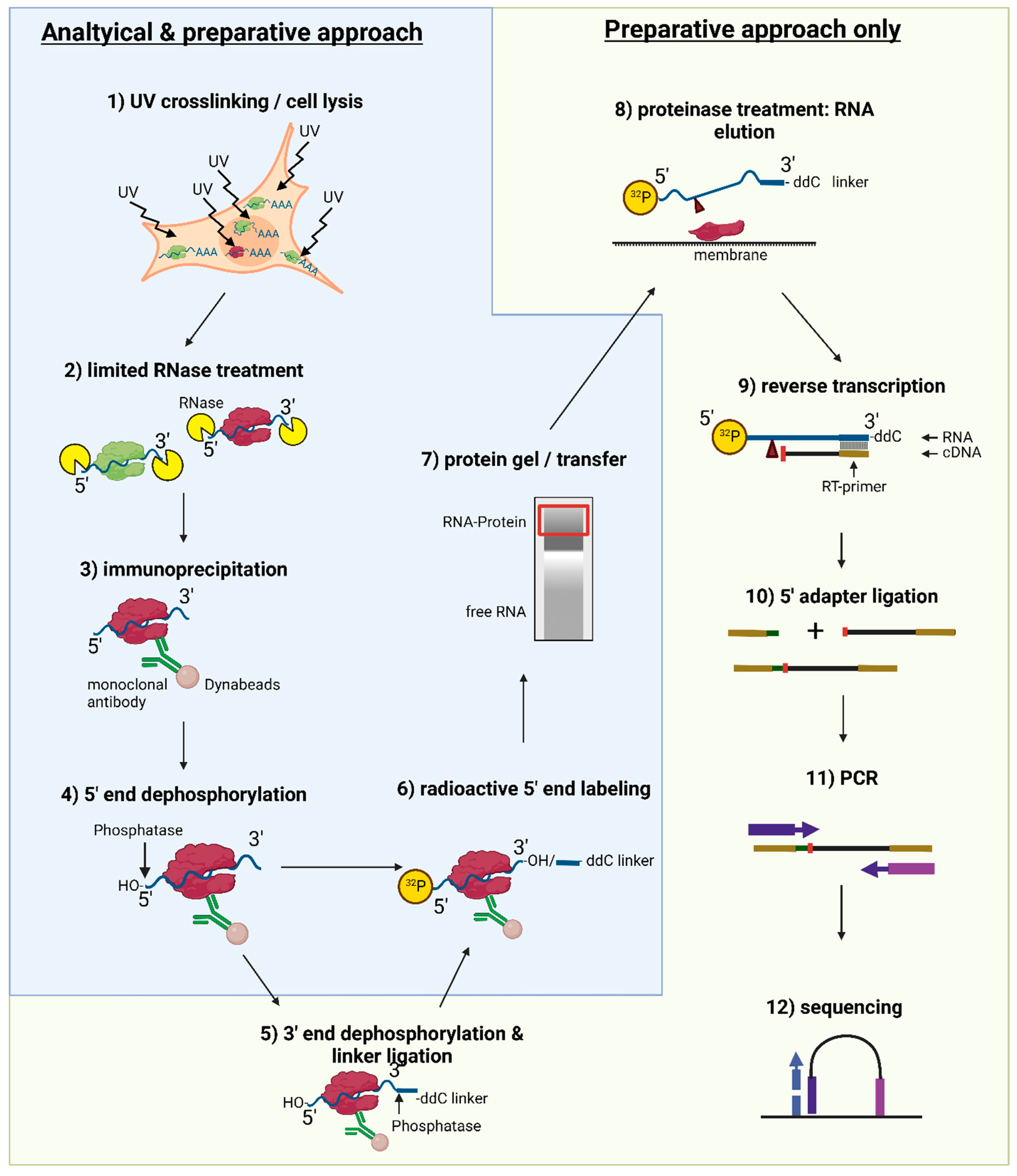
Global Approaches in Studying RNA-Binding Protein Interaction Networks: Trends in Biochemical Sciences
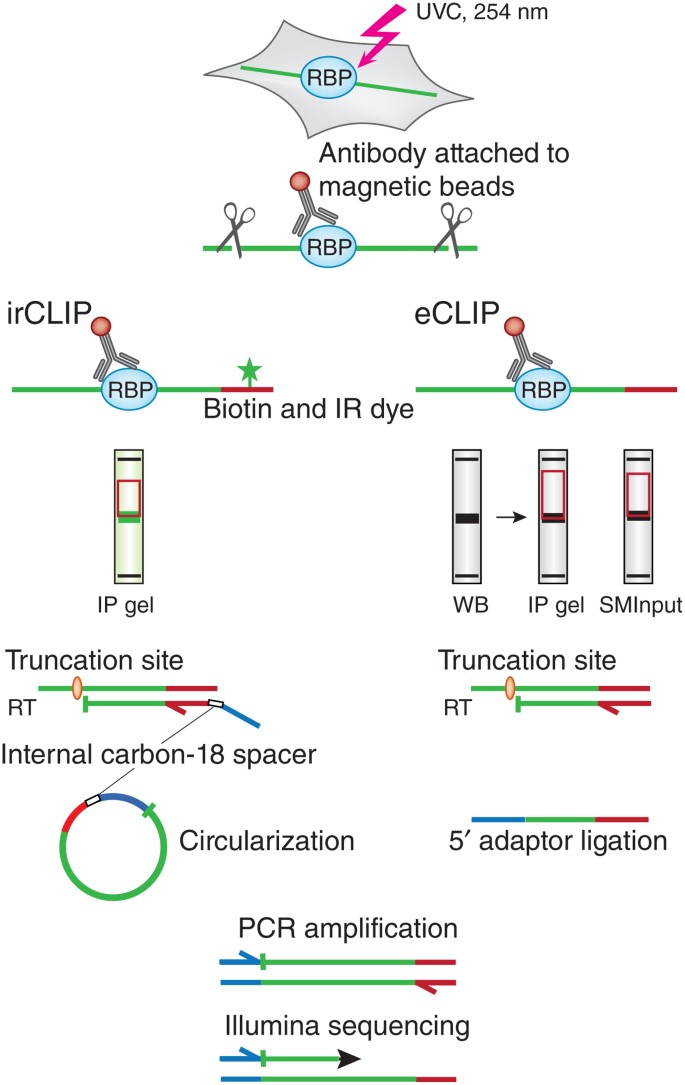
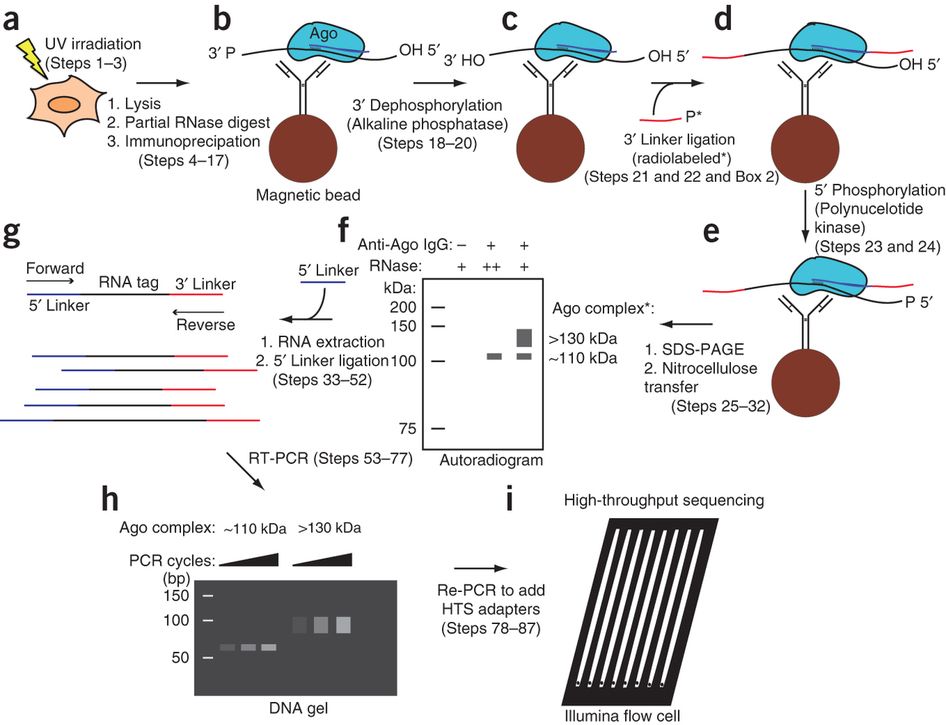
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein

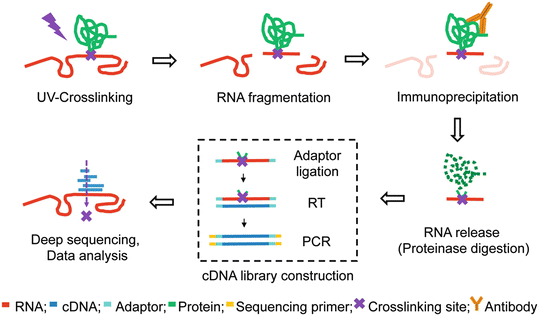
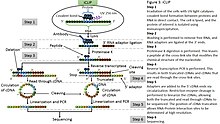

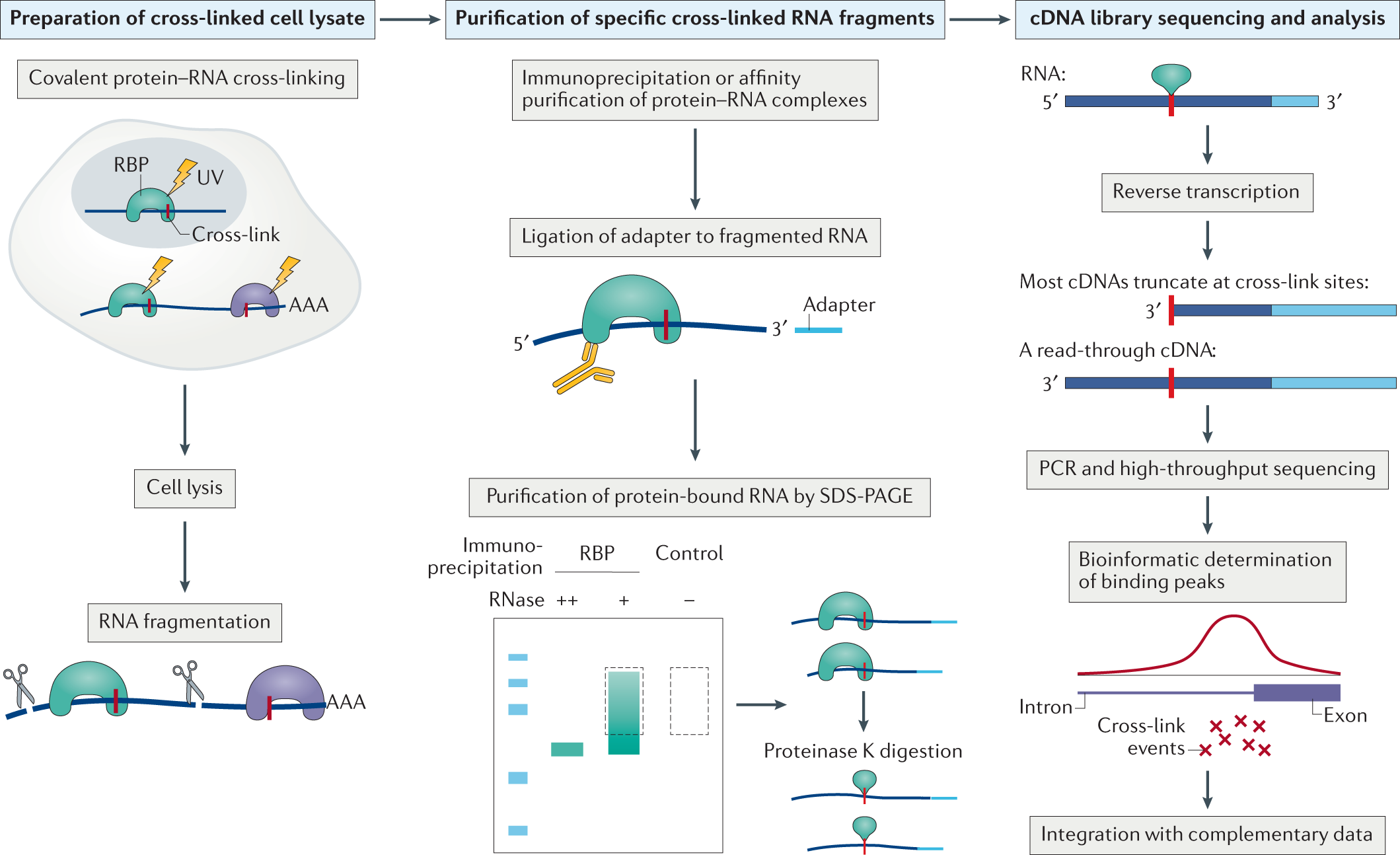
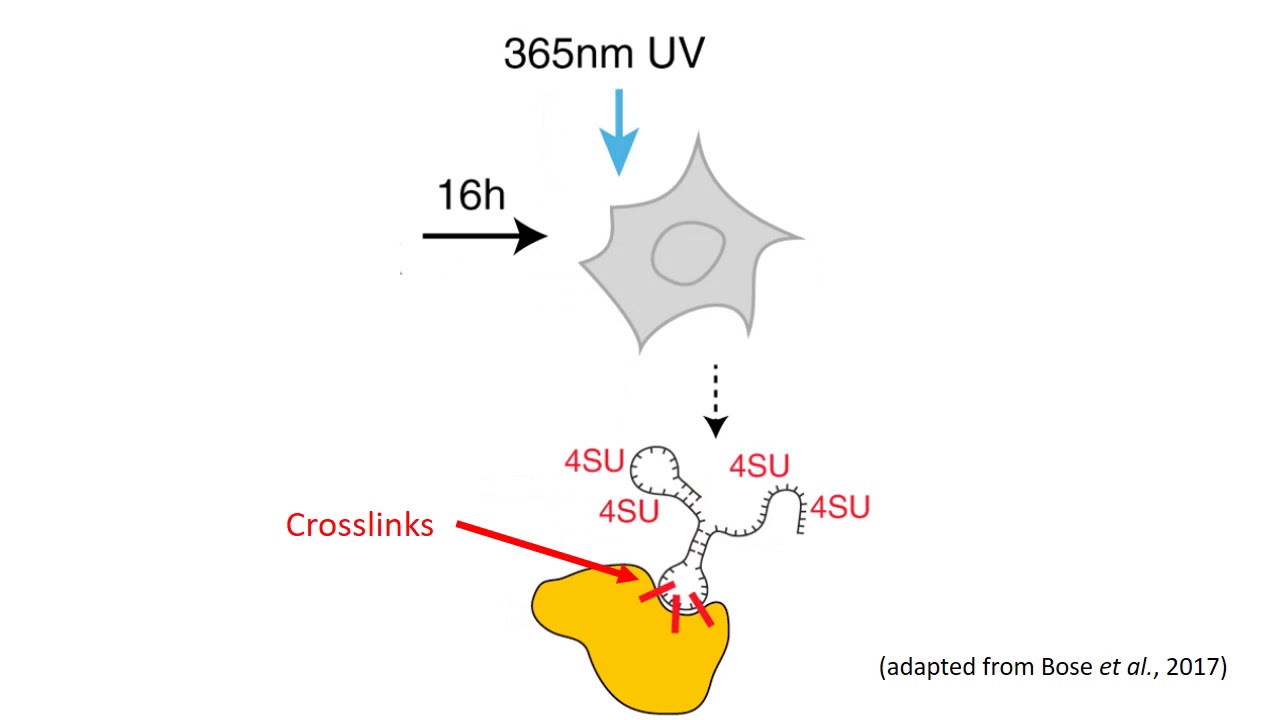

-2.jpg)